Researchers reveal conserved and specific regulation of poly(A) tail length in plants
DATE:2022-09-07
Poly(A) tail is a hallmark of eukaryotic mRNA, and its length plays an essential role in regulating mRNA metabolism. The length of the poly(A) tail is dynamically regulated by poly(A) polymerase and deadenylase. Recent progress in long-read sequencing platforms such as PacBio and Nanopore have enabled the development of methods that detect full-length mRNA with poly(A) tail information. However, a comprehensive resource for plant poly(A) tail length has yet to be established.
Associate Professor Jixian Zhai’s team from the Institute of Plant and Food Science at the Southern University of Science and Technology (SUSTech) recently published their research results as they applied single-molecule Nanopore sequencing and obtained over 120 million full-length RNAs with the poly(A) tail length information from various tissues and plant species. This dataset enables the genome-wide characterization of plant poly(A) tails at a single gene level across different tissues and species.
Their paper, entitled “An atlas of plant full-length RNA reveals tissue-specific and monocots-dicots conserved regulation of poly(A) tail length,” was published in Nature Plants, a high-impact academic journal covering a full range of disciplines concerned with plants.
The researchers previously developed a long-read-based method named FLEP-seq (Full-Length Elongating and Polyadenylated RNA sequencing) for characterizing nascent RNA. Here, they further adapted this method to study total RNA as an unbiased, poly(A)-enrichment-free measurement (FLEP-seq2) of full-length RNA with poly(A) tail information in plants.
The comprehensive atlas includes samples of seven different Arabidopsis tissues (seedling, root, shoot, leaf, inflorescence, seed, and pollen), as well as the shoot tissues of maize, rice, and soybean, each with two biological replicates (Figure 1). In total, they have constructed 20 FLEP-seq2 libraries that yielded ~276 million raw reads and ~121 million poly(A)+ reads, covering more than 10,000 genes with at least 20 reads in most libraries.
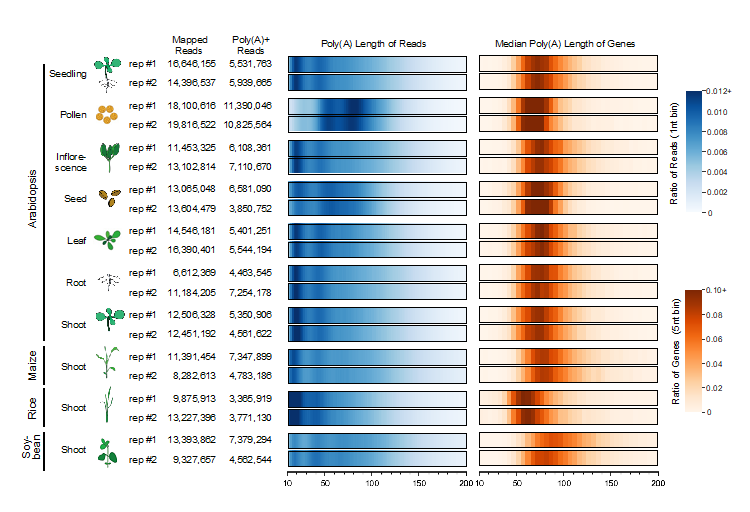
Figure 1. An atlas of plant poly(A) tail length
Prof. Zhai’s team found that the size of poly(A) tails shows peaks at ~20 and ~45 nt, presumably the sizes protected by one and two poly(A) binding proteins (PABP), respectively. However, the poly(A) tails in pollen and seed, tissues that may store mRNA for later uses, exhibit a distinct pattern with strong peaks at longer sizes (Figure 1). The data also revealed gene-specific regulation of poly(A) tail length, and mRNAs with the most prolonged half-lives have poly(A) tails that peak at ~45 nt, while the short-lived mRNAs have few tails in the corresponding range (Figure 2), suggesting that protection of poly(A) tail in this size by PABP is essential for mRNA stability.
In all four plant species tested, poly(A) tails in the nucleus are considerably longer than in the cytoplasm, implying a conserved rapid shortening process of poly(A) tail occurs before the mRNA is stabilized in the cytoplasm. Across species, the poly(A) tail lengths of orthologous genes are relatively conserved among different plant species, and thus might be selected under evolutionary pressure.
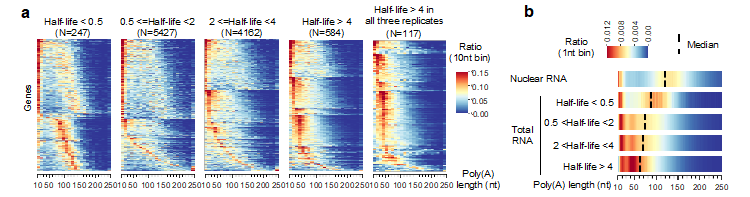
Figure 2. The poly(A) tail length distribution of genes with different mRNA half-lives. a) Heatmap plot showing the poly(A) tail length distribution of genes with different mRNA half-lives. Each row in the plot represents one gene. b) The bulk poly(A) tail length distribution of genes with different mRNA half-lives.
This study established a comprehensive dataset to facilitate future functional studies on how poly(A) tail length is influenced or regulated by various cis and trans factors in plants.
Dr. Jinbu Jia, Research Assistant Professor at SUSTech, and Wenqin Lu, a postdoctoral fellow of Assoc. Prof. Jixian Zhai’s group, are the co-first authors of this paper. Prof. Zhai is the corresponding author. Additional contributions were provided by Shanxi University.
The study was supported by the National Key R&D Program of China, National Natural Science Foundation of China (NSFC), Program for Guangdong Introducing Innovative and Entrepreneurial Teams, Shenzhen Sci-Tech Fund, and the Key Laboratory of Molecular Design for Plant Cell Factory of Guangdong Higher Education Institutes.
Paper link: https://www.nature.com/articles/s41477-022-01224-9
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
latest news
-
Dynamic changes in transposable elements shape human three-germ-layer differentiation
Date:2025-09-04
-
Researchers collaborate to uncover how SOD1 protects lysosome through autophagy
Date:2025-08-26
-
Researchers find 5-IP7 disrupts intestinal epithelial barrier and drives inflammation-induced colorectal cancer
Date:2025-08-26
-
Researchers decode molecular architecture and inhibition mechanism of human taurine transporter
Date:2025-08-22
