Dynamic changes in transposable elements shape human three-germ-layer differentiation
DATE:2025-09-04
Nearly half of the human genome is composed of transposable elements (TEs), previously regarded as “genomic junk” or potential genome destabilizing factors. Although abnormal activation of TEs has been well documented in embryonic stem cells and tumors, their dynamic changes and functional roles during a critical stage of human development have not been systematically explored. This knowledge gap has limited our deeper understanding of molecular developmental mechanisms and impeded research into developmental disorders.

A research group led by Associate Professor Andrew P. Hutchins from the Department of Systems Biology, School of Life Sciences at the Southern University of Science and Technology (SUSTech) has utilized long- and short-read RNA sequencing data to comprehensively analyze the dynamic expression, subcellular localization, and stability of TE transcripts during human germ-layer differentiation into three germ layers: endoderm, mesoderm and ectoderm. The findings not only reveal hierarchical and lineage-specific regulatory patterns of TEs but also provide new perspectives on the gene regulatory networks underpinning early human development.
Their paper, entitled “Transposable element expression and sub-cellular dynamics during hPSC differentiation to endoderm, mesoderm, and ectoderm lineages,” has been published in the journal Nature Communications.
The researchers showed that TEs exhibit striking lineage-specific and hierarchical differences across germ layers. For example, LTR6A is markedly upregulated in endoderm but strongly repressed in mesoderm and ectoderm, indicating that its transcriptional activity is finely tuned by cell fate decisions.
A particularly novel finding is the discovery of “TE switching.” Within the same TE family, TEs are not fixed to a single transcript but are incorporated into different transcriptional frameworks depending on cell type. For instance, in endoderm, certain LTRs serve as promoters driving neighboring gene expression, while in ectoderm, they are more often embedded within long non-coding RNAs. This “switching” mechanism reveals tissue-specific repurposing of TEs and suggests that they may function as alternative promoters, enhancers, or splice sites, thereby shaping lineage-specific gene regulatory networks.
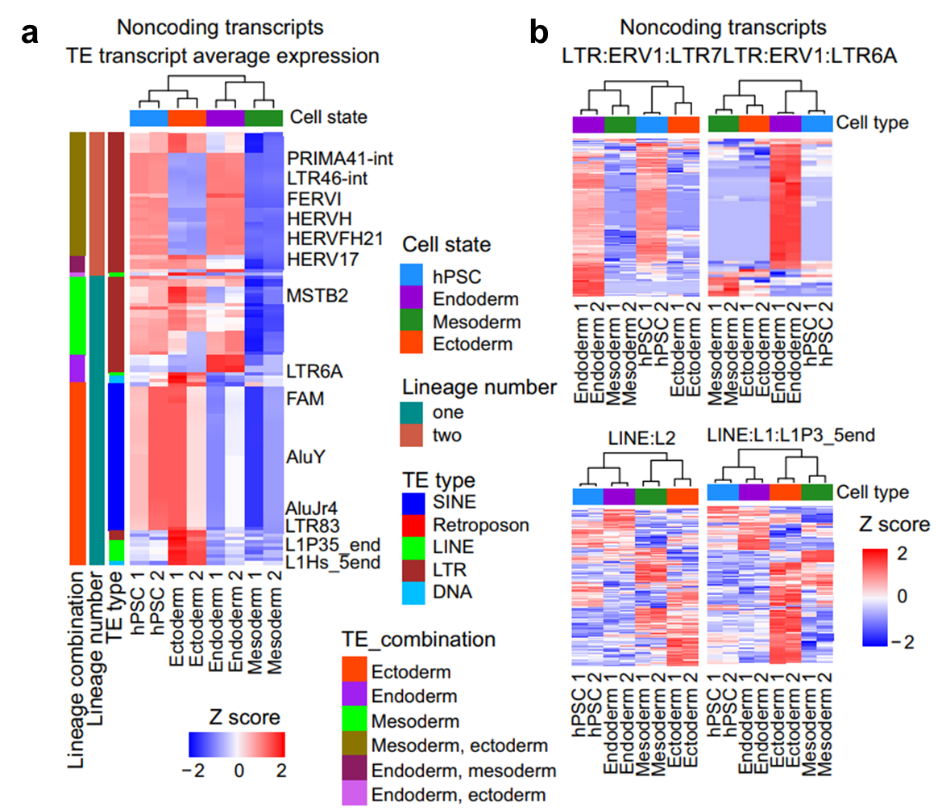
Figure 1. Lineage-specific expression of TE-associated non-coding transcripts. (a) Heatmap of overall expression changes in TE-related non-coding transcripts. (b) Heatmap of cell state-specific differentially expressed TE-related non-coding transcripts.
Beyond expression differences, the group also reveal cell-type-dependent subcellular localization and stability of TE transcripts. In undifferentiated human embryonic stem cells, TE transcripts are primarily located in the nucleoplasm or weakly bound to chromatin, suggesting they are in a “non-active” or easily degradable state. Upon differentiation into germ layers—especially in endoderm and mesoderm—TE transcripts increasingly anchor to nuclear chromatin, implying potential roles in modulating local chromatin accessibility or acting as regulatory platforms for gene expression.
Moreover, transcript stability differs among lineages: TE transcripts are more stable in mesoderm and ectoderm, supporting sustained functionality, but less stable in endoderm and undifferentiated cells, where they may function as transient regulatory signals. This reflects a differential utilization strategy whereby certain transcripts are stabilized to maintain cell fate while others are transiently activated to enable rapid developmental responses.
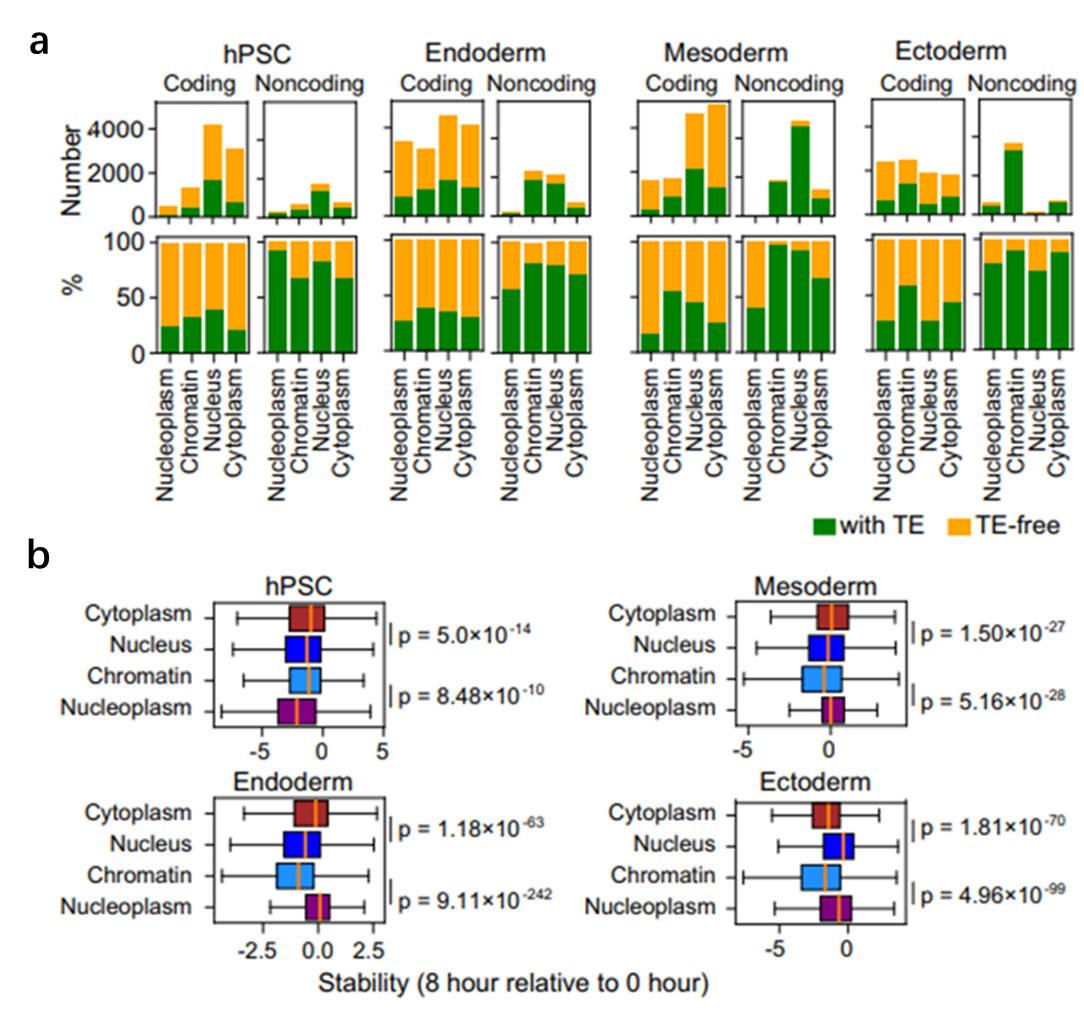
Figure 2. Subcellular localization and stability of TE transcripts. (a) Number and proportion of coding and non-coding transcripts localized to distinct subcellular compartments. (b) Stability of transcripts localized to different subcellular structures across cell states.
This study demonstrates that TEs are not merely “genomic parasites” but play important roles in early human embryonic development. It systematically uncovers the expression patterns, spatial distribution, and stability regulation of TEs across germ-layer differentiation, showing that TEs exert hierarchical and highly dynamic regulatory functions during lineage specification.
By establishing a solid molecular foundation for understanding TE function in development and disease, this work highlights their role as integral components of the genomic regulatory network. Further elucidation of these mechanisms may open new avenues for research and therapeutic strategies in regenerative medicine, neurodevelopmental disorders, and cancer.
Assistant Researcher Isaac A. Babarinde from the Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences (formerly of SUSTech), postdoctoral fellow Xiuling Fu from Columbia University, and Associate Professor Gang Ma from Southern Medical University (doctoral graduate of SUSTech) are co-first authors of the paper. Associate Professor Andrew P. Hutchins is the corresponding author, with SUSTech as the primary affiliated institution.
Paper link: https://www.nature.com/articles/s41467-025-63080-3
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
latest news
-
SUSTech Researchers Uncover the Molecular Mechanism of C1ql1/BAI3 Assemblies in CF-PC Synapse Development
Date:2025-12-29
-
Researchers discover mechanisms underlying how gravity competes with vision to guide brain’s inner compass
Date:2025-09-22
-
Dynamic changes in transposable elements shape human three-germ-layer differentiation
Date:2025-09-04
-
Researchers collaborate to uncover how SOD1 protects lysosome through autophagy
Date:2025-08-26
