Researchers analyze cryo-EM structures of human surfactant lipid transporter ABCA3
DATE:2022-04-13
Pulmonary surfactant, a mixture of approximately 90% lipids and 10% proteins, is secreted by the alveolar type II (AT2) epithelial cells and is dedicated to maintaining normal breathing by reducing alveolar surface tension. Prior to secretion, the pulmonary surfactant is stored in AT2 cell-specific, multi-lamellated lysosome-derived organelles named lamellar bodies (LBs). ABCA3 is localized in the limiting membrane of LBs and participates in surfactant biogenesis by transporting surfactant lipids, such as phosphatidylcholine (PC), phosphatidylglycerol (PG), and cholesterol, into LBs.
Mutations in the ABCA3 gene may cause surfactant deficiency, which leads to various lung disorders, such as fatal neonatal surfactant deficiency, chronic interstitial lung disease, and diffuse parenchymal lung disease. To date, no specific therapies exist for the hereditary disorders caused by ABCA3 mutations. Three-dimensional structural information of ABCA3 would help rationalize the disease-causing mutations with the final aim of developing therapies for patients.
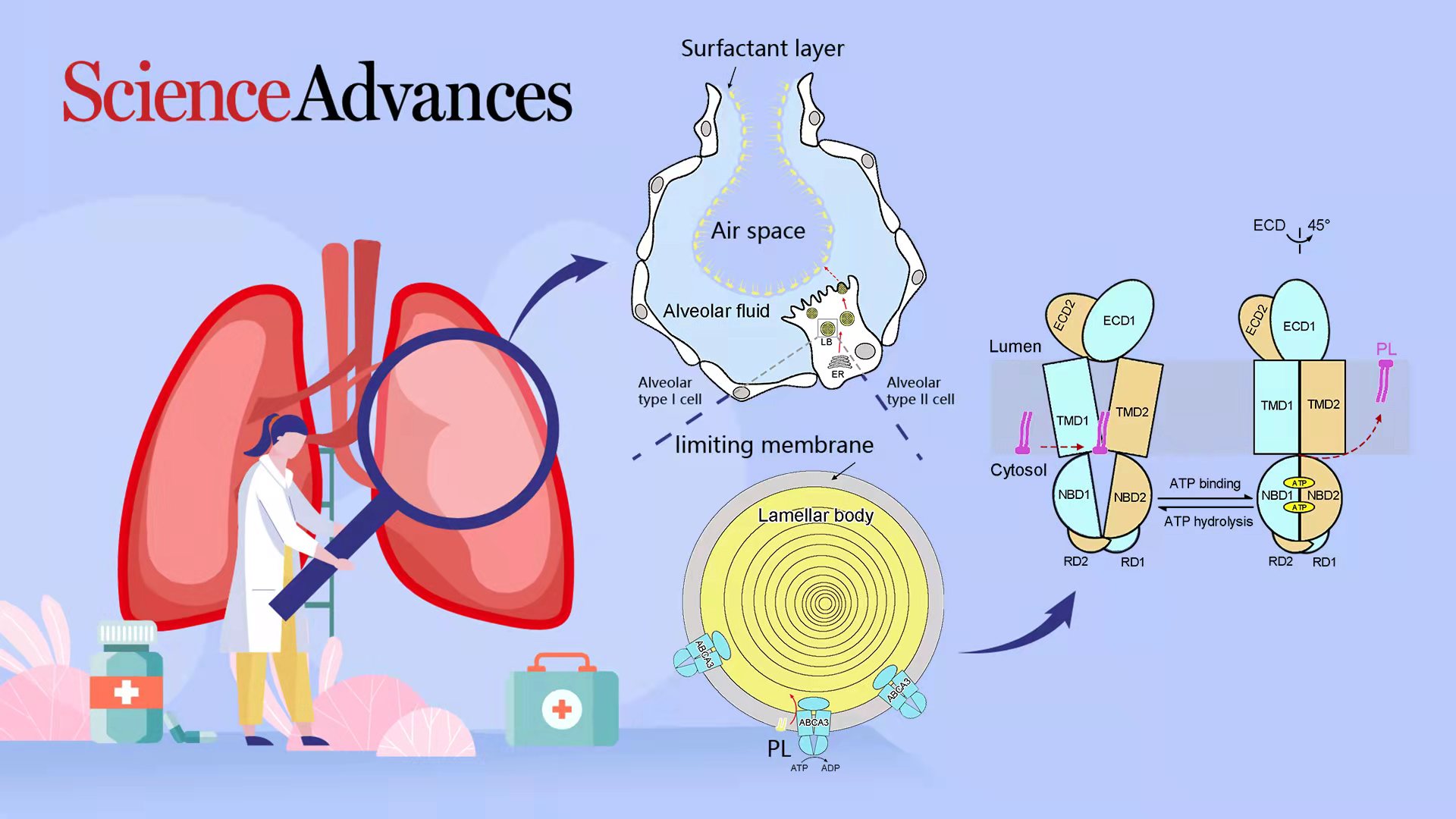
Professor Xin Gong’s research team from the School of Life Science at the Southern University of Science and Technology (SUSTech) recently published their findings on the cryo-EM structures of human ABCA3 in two distinct conformations, both at a resolution of 3.3 Å. Combined with biochemical data, they highlighted the substrate-binding site and features for proteolytic cleavage of ABCA3.
Their study, entitled “Cryo-EM structures of the human surfactant lipid transporter ABCA3,” was published in Science Advances, a multidisciplinary journal publishing impactful research in all areas of science. Their research also provided important information for rationalizing the disease-causing mutations in human ABCA3. It proposes a conserved ‘lateral access and extrusion’ mechanism for both lipid export and import mediated by ABCA transporters.
Their research presents purified human ABCA3 protein in different processed states, including pre-cleaved and mature states (Fig. 1 (D-E)). To explore the function of proteolytic processing, the ATPase activities of mutant proteins designed to mimic the pure unprocessed and processed ABCA3 indicate that the proteolytic processing of ABCA3 would not allosterically regulate the ATP hydrolysis (Fig. 1 (F-G)).
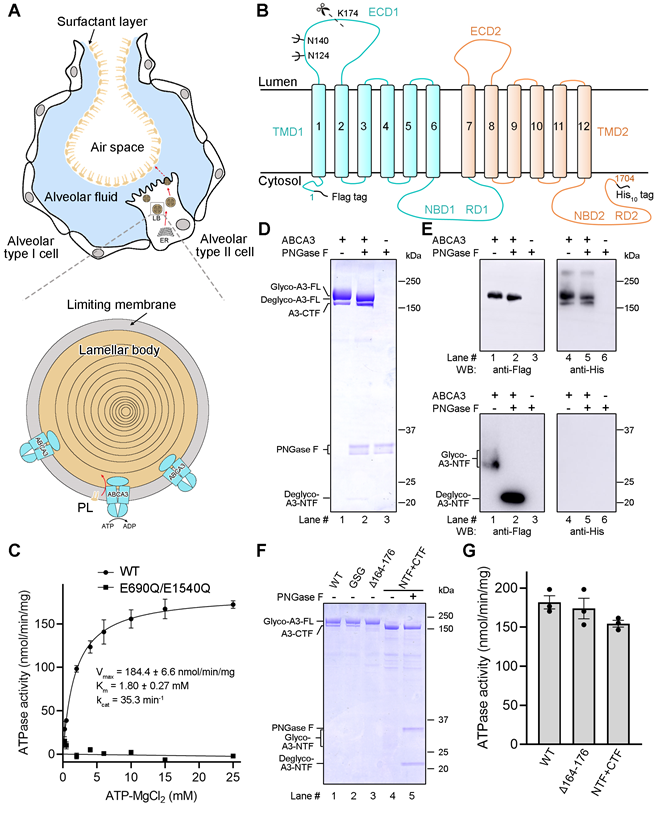
Figure 1. Biochemical characterization of human ABCA3
They further solved the cryo-EM structures of ABCA3 in both apo and ATP-binding conformations at the resolution of 3.3 Å (Fig. 2). These structures revealed two positively charged cavities within the TMDs as the potential substrate binding sites and delineate the ATP-dependent conformational changes to export the lipid substrates across the membrane. This suggests a conserved lipid transport mechanism for both lipid export and import mediated by ABCA transporters (Fig. 3). Together, this study provides important information for developing a therapeutic strategy targeted at ABCA3 in the future.
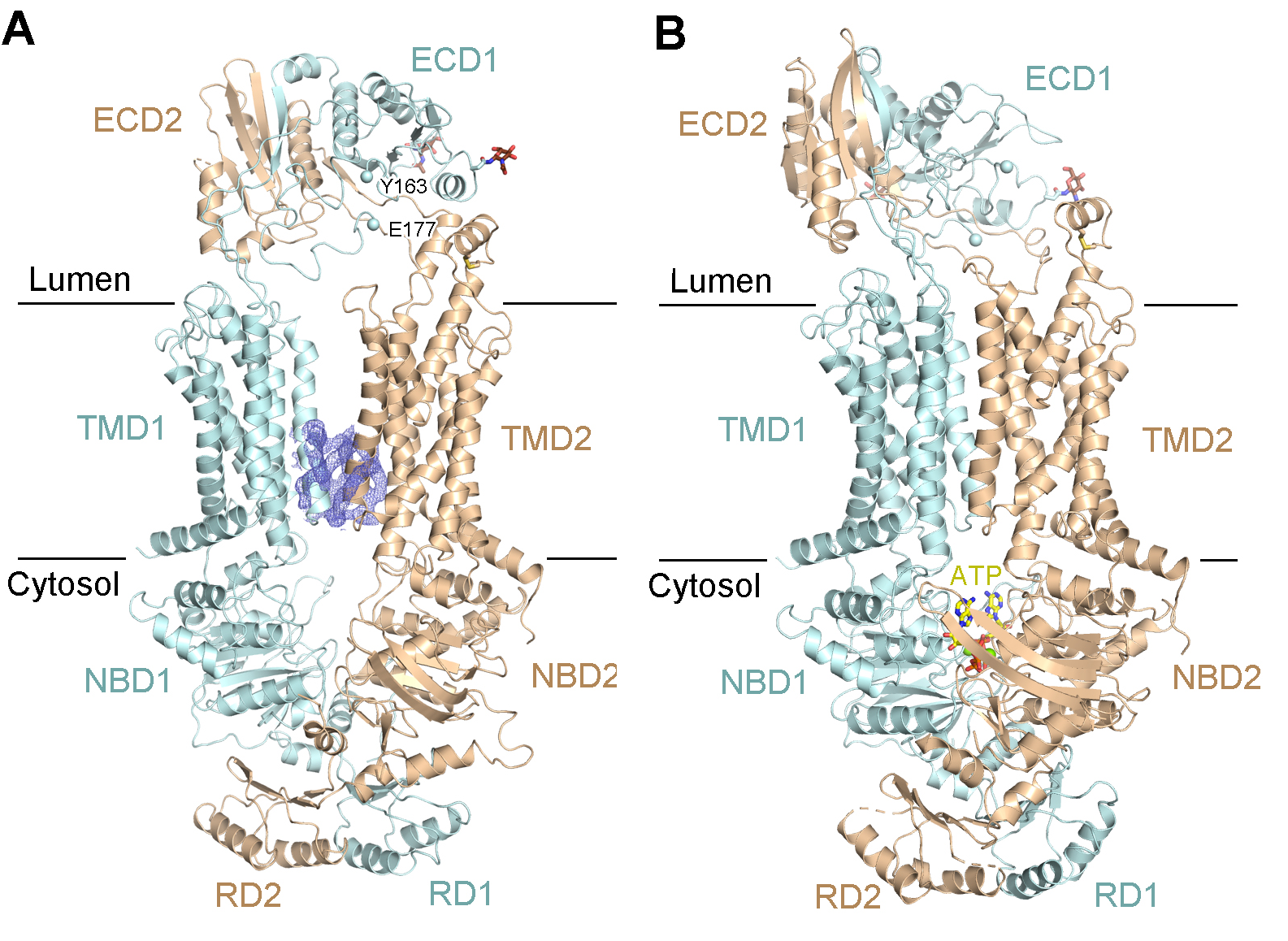
Figure 2. Cryo-EM structures of human ABCA3 in apo and ATP-binding states
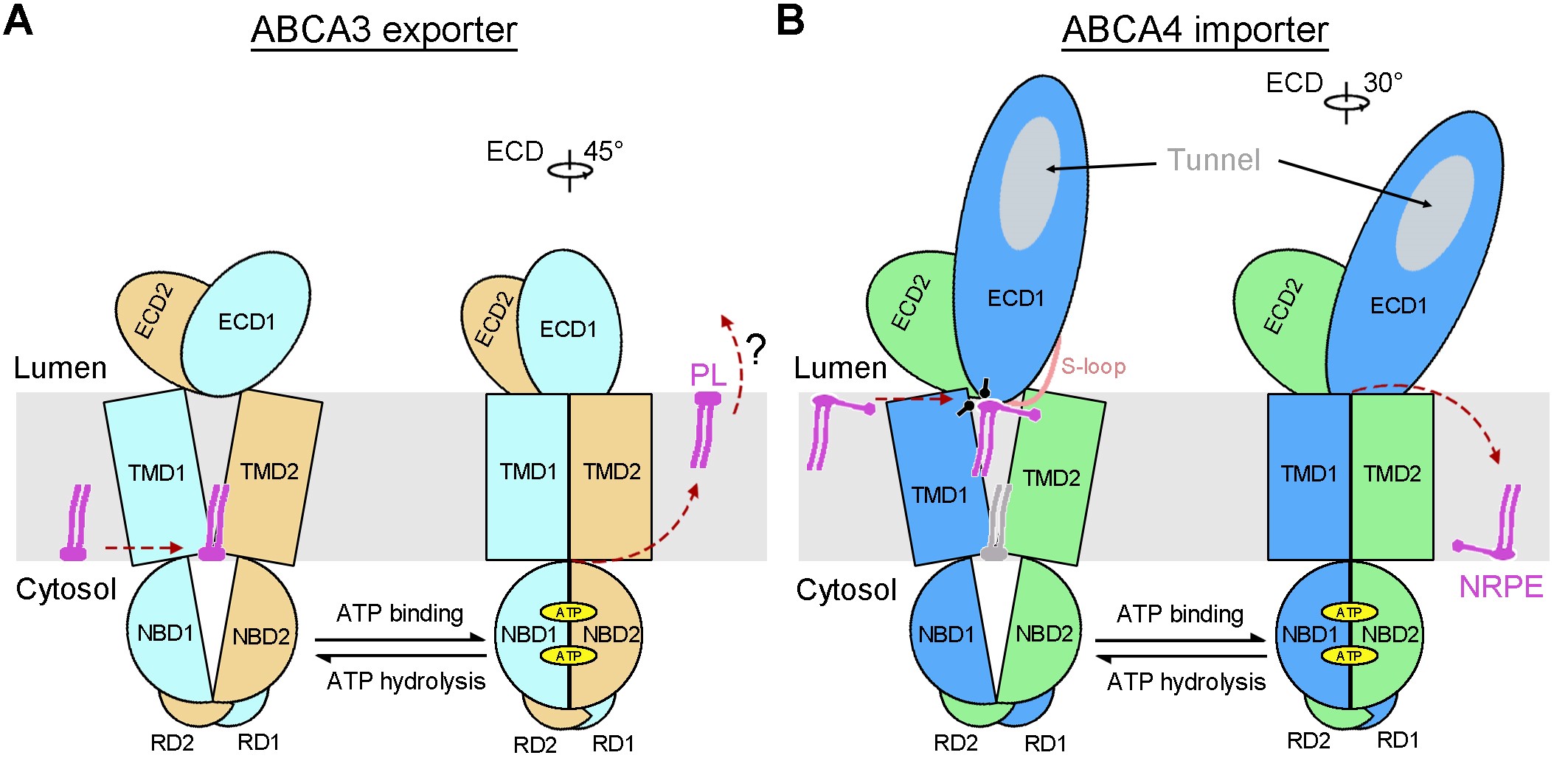
Figure 3. Working models of the ABCA transporters
This is another significant advance in the ABCA family of lipid transporters, following the publication of ABCA1 structure in Cell in 2017 and ABCA4 structure in Nature Communications in 2021 by Prof. Xin Gong.
Prof. Xin Gong from the School of Life Science at SUSTech is the corresponding author. Tian Xie, Zike Zhang, and Jian Yue, members of Prof. Gong’s research team, contributed equally to this study.
This research was supported by the National Natural Science Foundation of China (NSFC), the Natural Science Foundation of Guangdong Province for Distinguished Young Scientists, and the Shenzhen Science and Technology Program. Additionally, this work was assisted by the Cryo-Electron Microscopy Center of SUSTech.
Paper link: https://www.science.org/doi/10.1126/sciadv.abn3727
To read all stories about SUSTech science, subscribe to the monthly SUSTech Newsletter.
latest news
-
Dynamic changes in transposable elements shape human three-germ-layer differentiation
Date:2025-09-04
-
Researchers collaborate to uncover how SOD1 protects lysosome through autophagy
Date:2025-08-26
-
Researchers find 5-IP7 disrupts intestinal epithelial barrier and drives inflammation-induced colorectal cancer
Date:2025-08-26
-
Researchers decode molecular architecture and inhibition mechanism of human taurine transporter
Date:2025-08-22
